An integrative ENCODE resource for cancer: interpreting non-coding mutations and gene regulation
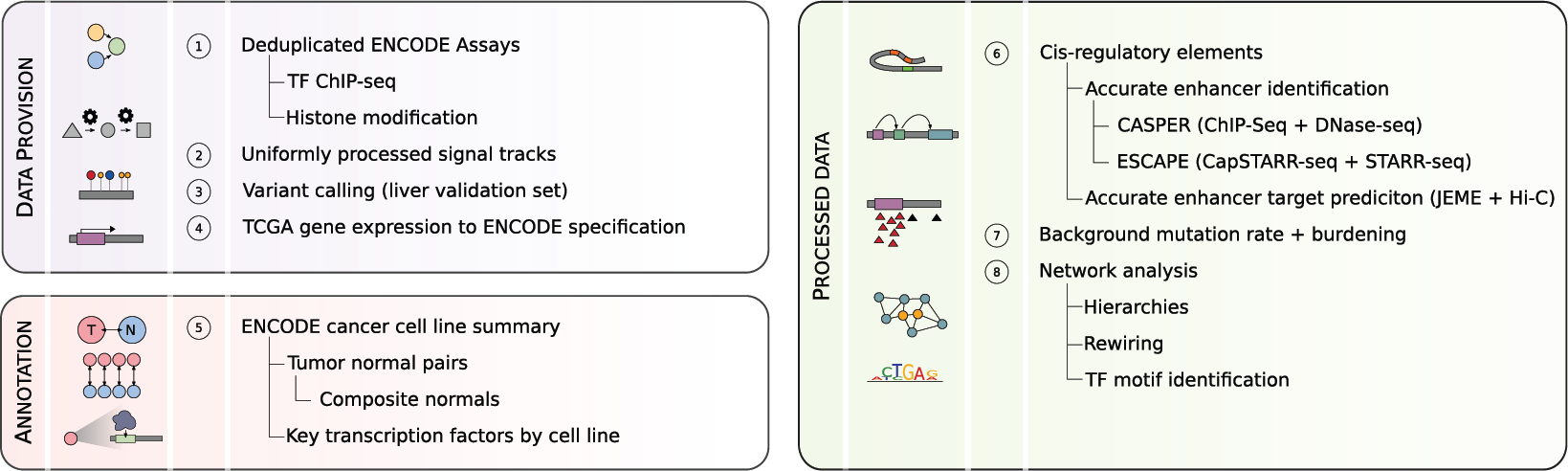
Supplementary Data
EN-CODEC.DATA
Summary of EN-CODEC experiments
| List of all EN-CODEC experiments (inc. external data) | EC-001-MET.encodec_all_experiments.csv |
| TF ChIP-seq data summary (Oct 2017 freeze) | EC-001-MET.encodec_Oct17_TF_ChIP-seq_data.csv |
| Deduplicated ChIP-seq list (Oct 2017 freeze) | EC-001-MET.encodec_Oct17_deduplicated_ChIP-seq_list.csv |
| Decuplicated ChIP-seq matrix (Oct 2017 freeze) | EC-001-MET.encodec_Oct17_deduplicated_ChIP-seq_matrix.csv |
| ENCODE key cell line ChIP-seq summary | EC-001-MET.encode_key_cell_line_ChIP-seq_data.csv |
Expression and variant calls
| Normalized cancer patient expression data | EC-001-COH.TCGA_data_2016_01_28.tar.gz |
| LIHC Germline | Available soon |
| LIHC Somatic | Available soon |
EN-CODEC.ANNOTATION
ENCODE cancer annotations
| ENCODE cancer cell type summary | EC-002-MET.encode_cancer_cell_type_summary.csv |
| ENCODE cancer cell line summary | EC-002-MET.encode_cancer_cell_line_summary.csv |
TF and RBP annotations
| K562 vs GM12878 common TF classification | EC-002-MET.encodec_KG_common_TF_class.csv |
| K562 vs GM12878 common TF summary | EC-002-MET.encodec_KG_common_TF_summary.csv |
Extended genes
| HepG2 extended gene definition | EC-002-EXT.merge.post.extendedGene.HepG2.txt |
| K562 extended gene definition | EC-002-EXT.merge.post.extendedGene.K562.txt |
| MCF-7 extended gene definition | EC-002-EXT.merge.post.extendedGene.MCF-7.txt |
EN-CODEC.ANALYSIS
MatchedFilter enhancer calls, SVM filtered
| GM12878 MatchedFilter enhancers | EC-003-CRE.MatchedFilter_GM12878.bed |
| GM12878 MatchedFilter enhancers merged | EC-003-CRE.MatchedFilter-merged_GM12878.bed |
| HepG2 MatchedFilter enhancers | EC-003-CRE.MatchedFilter_HepG2.bed |
| HepG2 MatchedFilter enhancers merged | EC-003-CRE.MatchedFilter-merged_HepG2.bed |
| K562 MatchedFilter enhancers | EC-003-CRE.MatchedFilter_K562.bed |
| K562 MatchedFilter enhancers merged | EC-003-CRE.MatchedFilter-merged_K562.bed |
| MCF-7 MatchedFilter enhancers | EC-003-CRE.MatchedFilter_MCF-7.bed |
| MCF-7 MatchedFilter enhancers merged | EC-003-CRE.MatchedFilter-merged_MCF-7.bed |
| A549 MatchedFilter enhancers | EC-003-CRE.MatchedFilter_A549.bed |
| A549 MatchedFilter enhancers merged | EC-003-CRE.MatchedFilter-merged_A549.bed |
| HeLa-S3 MatchedFilter enhancers | Available soon |
| HeLa-S3 MatchedFilter enhancers merged | Available soon |
| H1-hESC MatchedFilter enhancers | Available soon |
| H1-hESC MatchedFilter enhancers merged | Available soon |
ESCAPE enhancer peak calls
| GM12878 ESCAPE peak calls (raw) | Available soon |
| K562 ESCAPE peak calls (raw) | Available soon |
| MCF-7 ESCAPE peak calls (raw) | EC-003-CRE.ESCAPE_MCF-7.bed |
| LNCaP ESCAPE peak calls (raw) | EC-003-CRE.ESCAPE_LNCaP.bed |
| GM12878 ESCAPE peak calls (lenient, FC > 1.2) | Available soon |
| K562 ESCAPE peak calls (lenient, FC > 1.2) | Available soon |
| MCF-7 ESCAPE peak calls (lenient, FC > 1.2) | EC-003-CRE.ESCAPE_MCF-7_lenient.bed |
| LNCaP ESCAPE peak calls (lenient, FC > 1.2) | EC-003-CRE.ESCAPE_LNCaP_lenient.bed |
| GM12878 ESCAPE peak calls (strict, FC > 2) | Available soon |
| K562 ESCAPE peak calls (strict, FC > 2) | Available soon |
| MCF-7 ESCAPE peak calls (strict, FC > 2) | EC-003-CRE.ESCAPE_MCF-7_strict.bed |
| LNCaP ESCAPE peak calls (strict, FC > 2) | EC-003-CRE.ESCAPE_LNCaP_strict.bed |
JEME enhancer target predictions
| GM12878 JEME enhancer target gene prediction | EC-003-CRE.JEME_of_MatchedFilter_GM12878.bed |
| HepG2 JEME enhancer target gene prediction | EC-003-CRE.JEME_of_MatchedFilter_HepG2.bed |
| K562 JEME enhancer target gene prediction | EC-003-CRE.JEME_of_MatchedFilter_K562.bed |
| MCF-7 JEME enhancer target gene prediction | EC-003-CRE.JEME_of_MatchedFilter_MCF-7.bed |
| A549 JEME enhancer target gene prediction | Available soon |
| HeLa-S3 JEME enhancer target gene prediction | Available soon |
| H1-hESC JEME enhancer target gene prediction | Available soon |
Integrative enhancer annotations with accession to the Encyclopedia cRE
| GM12878 MatchedFilterxESCAPE integrated annotation (with mapping to cRE accession) | EC-003-CRE.MatchedFilterxESCAPE_GM12878.bed |
| K562 MatchedFilterxESCAPE integrated annotation (with mapping to cRE accession) | EC-003-CRE.MatchedFilterxESCAPE_K562.bed |
| GM12878 MatchedFilterxESCAPExJEME integrative enhancer-target gene annotation | EC-003-CRE.MatchedFilterxESCAPExJEME-targetGene_GM12878.bed |
| K562 MatchedFilterxESCAPExJEME integrative enhancer-target gene annotation | EC-003-CRE.MatchedFilterxESCAPExJEME-targetGene_K562.bed |
| GM12878 MatchedFilterxESCAPExJEME integrative enhancer-target gene annotation, Hi-C annotated (last col) | EC-003-CRE.MatchedFilterxESCAPExJEME-targetGene_FitHiC_GM12878.bed |
| K562 MatchedFilterxESCAPExJEME integrative enhancer-target gene annotation, Hi-C annotated (last col) | EC-003-CRE.MatchedFilterxESCAPExJEME-targetGene_FitHiC_K562.bed |
| GM12878 MatchedFilterxESCAPExJEME integrative enhancer-target gene annotation, Hi-C annotated, merged | EC-003-CRE.MatchedFilterxESCAPExJEME-targetGene_FitHiC_GM12878_merged.bed |
| K562 MatchedFilterxESCAPExJEME integrative enhancer-target gene annotation, Hi-C annotated, merged | EC-003-CRE.MatchedFilterxESCAPExJEME-targetGene_FitHiC_K562_merged.bed |
| GM12878 MatchedFilterxESCAPExJEME integrative enhancer-target gene annotation, Hi-C filtered (q-val<0.1) | EC-003-CRE.MatchedFilterxESCAPExJEME-targetGene_FitHiC-filtered_GM12878.bed |
| K562 MatchedFilterxESCAPExJEME integrative enhancer-target gene annotation, Hi-C filtered (q-val<0.1) | EC-003-CRE.MatchedFilterxESCAPExJEME-targetGene_FitHiC-filtered_K562.bed |
| GM12878 MatchedFilterxESCAPExJEME integrative enhancer-target gene annotation, Hi-C filtered, merged | EC-003-CRE.MatchedFilterxESCAPExJEME-targetGene_FitHiC-filtered_merged_GM12878.bed |
| K562 MatchedFilterxESCAPExJEME integrative enhancer-target gene annotation, Hi-C filtered, merged | EC-003-CRE.MatchedFilterxESCAPExJEME-targetGene_FitHiC-filtered_merged_K562.bed |
| GM12878 MatchedFilterxESCAPExJEMExcRE compact integrative enhancer-target gene annotation, Hi-C filtered, merged | EC-003-CRE.hg19-cRExMatchedFilterxJEME_Hi-C_GM12878_merged.bed |
| K562 MatchedFilterxESCAPExJEMExcRE compact integrative enhancer-target gene annotation, Hi-C filtered, merged | EC-003-CRE.hg19-cRExMatchedFilterxJEME_Hi-C_K562_merged.bed |
| GM12878 MatchedFilterxESCAPExJEMExcRE compact integrative enhancer-target gene annotation, Hi-C filtered, merged, trimmed by TFBS | EC-003-CRE.hg19-cRExMatchedFilterxJEME_Hi-C_GM12878_merged_trimmed.bed |
| K562 MatchedFilterxESCAPExJEMExcRE compact integrative enhancer-target gene annotation, Hi-C filtered, merged, trimmed by TFBS | EC-003-CRE.hg19-cRExMatchedFilterxJEME_Hi-C_K562_merged_trimmed.bed |
Extended genes
| HepG2 P-value package for extended genes | EC-003-EXT.pval_HepG2.tar.gz |
| K562 P-value package for extended genes | EC-003-EXT.pval_K562.tar.gz |
| MCF-7 P-value package for extended genes | EC-003-EXT.pval_MCF-7.tar.gz |
Background mutation rate (BMR)
| BMR detailed list of features | EC-003-BMR.feature_details.txt |
| Covariate matrix 100kb resolution | EC-003-BMR.covariate_region_100kb.txt |
| Covariate matrix 1mb resolution | EC-003-BMR.covariate_region_1mb.txt |
| Description of BMR estimation parameters | EC-003-BMR.README.txt |
| Blood cancer BMR estimation parameter | EC-003-BMR.BloodCan_dispersion.txt |
| Blood cancer BMR estimation parameter | EC-003-BMR.BloodCan_mu_nb.txt |
| Blood cancer BMR estimation parameter | EC-003-BMR.BloodCan_mu_pois.txt |
| Blood cancer BMR estimation parameter | EC-003-BMR.BloodCan_theta_nb.txt |
| Breast cancer BMR estimation parameter | EC-003-BMR.BreastCan_dispersion.txt |
| Breast cancer BMR estimation parameter | EC-003-BMR.BreastCan_mu_nb.txt |
| Breast cancer BMR estimation parameter | EC-003-BMR.BreastCan_mu_pois.txt |
| Breast cancer BMR estimation parameter | EC-003-BMR.BreastCan_theta_nb.txt |
| Liver cancer BMR estimation parameter | EC-003-BMR.LiverCan_dispersion.txt |
| Liver cancer BMR estimation parameter | EC-003-BMR.LiverCan_mu_nb.txt |
| Liver cancer BMR estimation parameter | EC-003-BMR.LiverCan_mu_pois.txt |
| Liver cancer BMR estimation parameter | EC-003-BMR.LiverCan_theta_nb.txt |
| Lung cancer BMR estimation parameter | EC-003-BMR.LungCan_dispersion.txt |
| Lung cancer BMR estimation parameter | EC-003-BMR.LungCan_mu_nb.txt |
| Lung cancer BMR estimation parameter | EC-003-BMR.LungCan_mu_pois.txt |
| Lung cancer BMR estimation parameter | EC-003-BMR.LungCan_theta_nb.txt |
Regulatory score
| TF regulatory score description | EC-003-DBP.regulatoryScore_README.txt |
| TF regulatory score package | EC-003-DBP.regulatoryScore.tar.gz |
| RBP regulatory score package | Available soon |
Cell-type-specific network
| TIP Profiles of ENCODE TF ChIP-seq experiments | EC-003-NET.ENCODE_TIP_Profiles_693.txt |
| GM12878 Proximal TIP-based network | EC-003-NET.edgeList_TIP_GM12878.tsv |
| K562 Proximal TIP-based network | EC-003-NET.edgeList_TIP_K562.tsv |
| GM12878 Proximal TSS-based network, using all TSS annotations | EC-003-NET.edgeList_TSS_GM12878.tsv |
| K562 Proximal TSS-based network, using all TSS annotations | EC-003-NET.edgeList_TSS_K562.tsv |
| GM12878 Distal enhancer-based network | EC-003-NET.edgeList_ENH_GM12878.tsv |
| K562 Distal enhancer-based network | EC-003-NET.edgeList_ENH_K562.tsv |
| A549 Proximal TSS-based network, using 1 TSS annotation per gene | EC-003-NET.edgeList_uniqTSS2500_A549.tsv |
| GM12878 Proximal TSS-based network, using 1 TSS annotation per gene | EC-003-NET.edgeList_uniqTSS2500_GM12878.tsv |
| H1-hESC Proximal TSS-based network, using 1 TSS annotation per gene | EC-003-NET.edgeList_uniqTSS2500_H1-hESC.tsv |
| HeLa-S3 Proximal TSS-based network, using 1 TSS annotation per gene | EC-003-NET.edgeList_uniqTSS2500_HeLa-S3.tsv |
| HepG2 Proximal TSS-based network, using 1 TSS annotation per gene | EC-003-NET.edgeList_uniqTSS2500_HepG2.tsv |
| IMR-90 Proximal TSS-based network, using 1 TSS annotation per gene | EC-003-NET.edgeList_uniqTSS2500_IMR-90.tsv |
| K562 Proximal TSS-based network, using 1 TSS annotation per gene | EC-003-NET.edgeList_uniqTSS2500_K562.tsv |
| MCF-10A Proximal TSS-based network, using 1 TSS annotation per gene | EC-003-NET.edgeList_uniqTSS2500_MCF-10A.tsv |
| MCF-7 Proximal TSS-based network, using 1 TSS annotation per gene | EC-003-NET.edgeList_uniqTSS2500_MCF-7.tsv |
| Liver Proximal TSS-based network, using 1 TSS annotation per gene | EC-003-NET.edgeList_uniqTSS2500_liver.tsv |
| Lung Proximal TSS-based network, using 1 TSS annotation per gene | EC-003-NET.edgeList_uniqTSS2500_lung.tsv |
Reconciled merged network
| Proximal TIP-based merged network | EC-003-NET.mergedNet_TIP_FDR_0.1.tsv |
| Proximal TSS-based merged network | EC-003-NET.mergedNet_uniqTSS2500_ALL.tsv |
| Probability-based network description | EC-003-NET.probNet_README.txt |
| Probability-based TF network | EC-003-NET.probNet_TF.txt |
| Probability-based RBP network | EC-003-NET.probNet_RBP.txt |
| Gene ID conversion table | EC-003-NET.Ensemble2EntrezGeneID.tsv |
Network hierachy
| Merged network hierachy description | EC-003-NET.hierarchy_mergedNet_README.txt |
| Merged network hierachy, TIP-based | EC-003-NET.hierarchy_mergedNet_TFss.merged.TIP.network.txt |
| Merged network hierachy, mean t-value | EC-003-NET.hierarchy_mergedNet_tval.mean.across.patients.txt |
| Cell type specific network hierachy documentation | EC-003-NET.hierarchy_cellTypeSpecNet_README.txt |
| Cell type specific network hierachy node properties | EC-003-NET.hierarchy_cellTypeSpecNet_node.properties.txt |
| GM12878 network hierachy | EC-003-NET.hierarchy_cellTypeSpecNet_GM12878.tfcom.ss.txt |
| K562 network hierachy | EC-003-NET.hierarchy_cellTypeSpecNet_K562.tfcom.ss.txt |
Network rewiring and rewiring index
| CML proximal-distal combined TF-GENE network rewiring, TIP-based | EC-003-NET.KG_TIP_ENH_mnet.csv |
| CML proximal-distal combined TF-GENE network rewiring, TSS-based | EC-003-NET.KG_TSS_ENH_mnet.csv |
| CML proximal-distal combined TF-TF network rewiring, TIP-based | EC-003-NET.KG_TIP_ENH_tfnet.csv |
| CML proximal-distal combined TF-TF network rewiring, TSS-based | EC-003-NET.KG_TSS_ENH_tfnet.csv |
| TF attributes, TIP-based | EC-003-NET.KG_TIP_ENH_TFattrib.csv |
| TF attributes, TSS-based | EC-003-NET.KG_TSS_ENH_TFattrib.csv |
| GENE attributes, TIP-based | EC-003-NET.KG_TIP_ENH_GENEattrib.csv |
| GENE attributes, TSS-based | EC-003-NET.KG_TSS_ENH_GENEattrib.csv |
| CML rewiring index, direct connection, TIP-based | EC-003-NET.KG_rewiringIndex_TIP+ENH.csv |
| CML rewiring index, direct connection, TSS-based | EC-003-NET.KG_rewiringIndex_TSS+ENH.csv |
| CML rewiring index, gene community-based | EC-003-NET.KG_rewiringIndex-geneCommunity_TFrand_mixedM_median.txt |
| CML rewiring index, gene community-based, cubic-rooted | EC-003-NET.KG_rewiringIndex-geneCommunity_TFrand_mixedM_median_cubicroot.txt |
EN-CODEC.VALIDATION
Experimental validations
| Luciferase CRE validation raw data | EC-004-CRE.luciferase_raw.csv |
| Luciferase CRE validation normalized data | EC-004-CRE.luciferase_normalized.csv |
| MYC knockdown experiment description | EC-004-DBP.README.txt |
| MYC siRNA control | EC-004-DBP.MYC-KD_geneFPKM_control.txt |
| MYC siRNA knockdown | EC-004-DBP.MYC-KD_geneFPKM_knockdown.txt |
EN-CODEC.SOFTWARE
Code releases
| BMR software package | Available soon |
| ESCAPE software package | Available soon |
| Rewiring direct connection software package | EC-005-NET.Rewiring_directConnection_related.tar.gz |
| Rewiring mixed membership software package | EC-005-NET.Rewiring_geneCommunity_related.tar.gz |